Study identifies regions on the SARS-CoV-2 genome linked to virus replication and transmission
Since COVID-19 emerged, scientists worldwide have sequenced more than 6 million genomes of SARS-CoV-2, the coronavirus behind the disease. Today, the number of SARS-CoV-2 genome sequences exceeds the total number of all other viral genomes.
Genomes are valuable because they provide a record of the evolution of a virus in ahuman host, along with information on the emergence of mutations. In a study, published in theJournal of Biosafety and Biosecurity, a team of researchers analyzed 2.8 million of the sequenced SARS-CoV-2 genomes and used the results to compile a 'mutations blacklist' of virus weak spots, and a 'whitelist' of mutations that make SARS-CoV-2 more transmissible. In addition, the team has developed an online SARS-CoV-2 mutation and variant monitoring and pre-warning system (MVMPS).
Professor Jianguo Xu of China's National Institute for Communicable Diseases Control and Prevention led the study. Reflecting on the findings, he explained that they "found that six types of gene-level mutations had reached saturation in the sequenced genome population we examined—when you reach saturation, it means that no novel mutations should appear in the future, even if more genomes are sequenced. This result points to most subsequent variants of SARS-CoV-2 being a combination of existing high-frequency mutations, rather than a newly-emerged mutation. For example, the variant BA.5 has a L452R mutation that escapes host immunity. Although this mutation appears in the Omicron variant, it was also found in the Delta variant. This kind of information is crucial, as it suggests these mutations might have anegative effecton viral survival, replication and transmission, information that can aid vaccine and drug development research." These mutations were used to compile the team's SARS-CoV-2 'mutations blacklist'.
He continued: "Some mutations can affect the virus transmission rate. We identified a total of 185 mutations that are significantly positively correlated with viral transmission, information that scientists can use to evaluate the transmissibility of new variants." These findings were used to create the team's SARS-CoV-2 'mutations whitelist'.
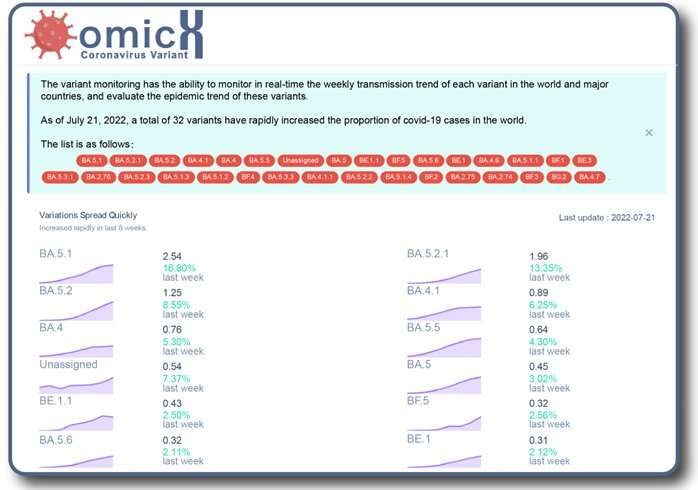
In addition, the team has built an online alert system—the MVMPS—which is home to a dynamic curve that shows, inreal-time, proportions of each mutation andvariantin all the SARS-CoV-2 strains. According to Prof. Xu, the hope is that the MVMPS will not only monitor the occurrence and development of thesemutationsand variants, but provide vital early warninginformationto help with prevention andcontrol measures.